4. Inside CGAR Report¶
For each sample, the generated report shows the name (identifier) of the chosen sample with 9 sections of variants organized by their analytical implications.
- (Restricted) Disease-associated variants reported in HGMD -
HGMD - Disease-associated variants reported in ClinVar -
ClinVar - (Restricted) Variants on genes associated with rare-diseases in OrphaData -
Orphanet - Variants on genes with putative association to user-defined phenotype -
Phenotype associated - Secondary findings -
ACMG59 - Variants with potential pharmacogenomic implications -
Pharmacogenomics - Variants on user-defined set of genes -
Focused report - De novo variant candidates from trio -
De novo candidates - (Somatic) Variants potentially associated with cancer -
Cancer
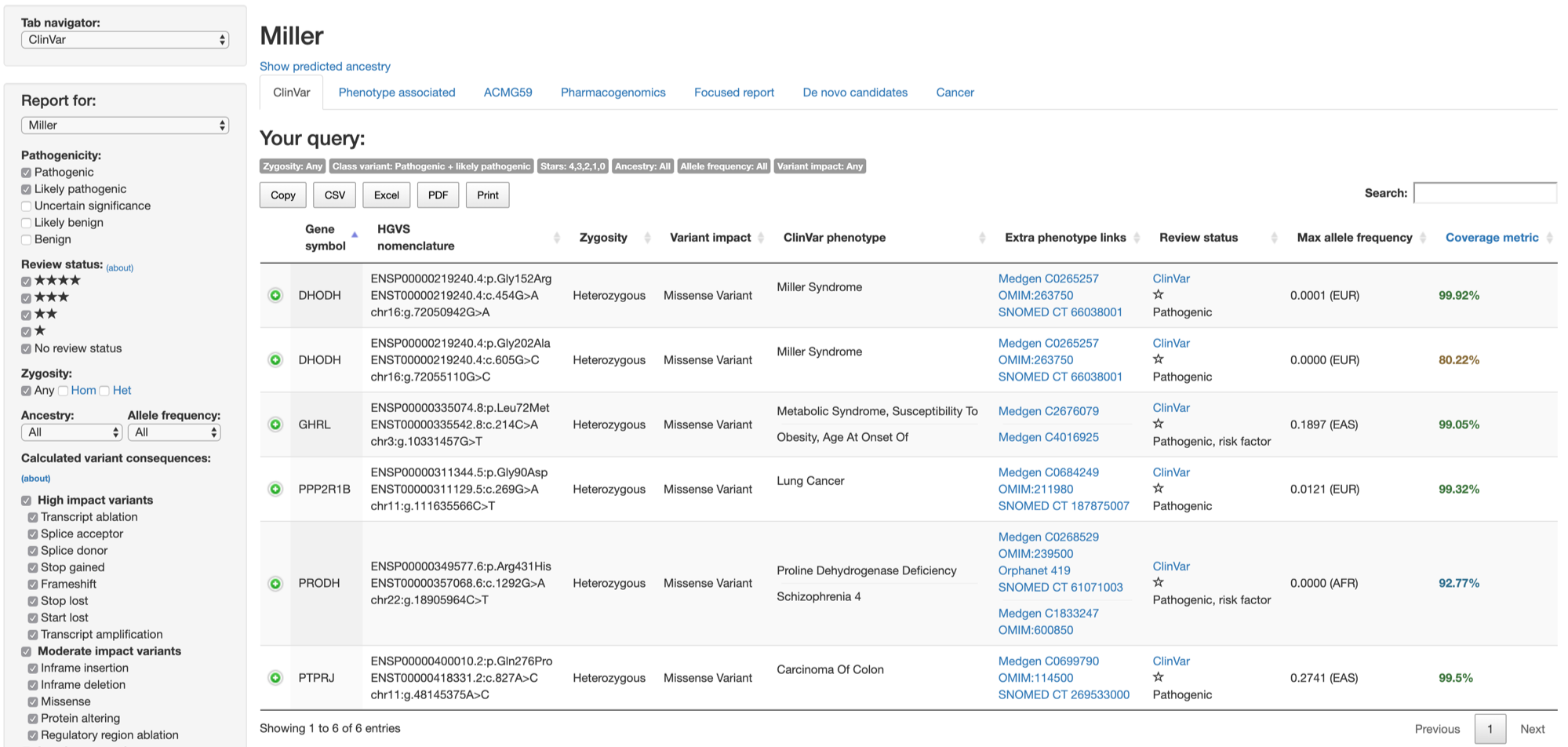
The side manu on the left reflects settings for current section and provides means to change settings and to reanalyze. The main table on the right presents the essential information on variants as well as links to further details or external resources.
In the following sections, each component and section in CGAR report is described in detail.
- 4.1. Genotype-based prediction of ancestral composition
- 4.2. Side menu and main table
- 4.3. HGMD variants
- 4.4. ClinVar variants
- 4.5. Genes associated with rare phenotype
- 4.6. Putative phenotype-associated genes
- 4.7. Secondary findings
- 4.8. Pharmacogenomic variants
- 4.9. User-defined set of genes
- 4.10. De novo variant candidates
- 4.11. Cancer-associated genes and variants