4.3. HGMD variants¶
Warning
The contents for this section is available only for users licensed from the Human Gene Mutation Database (HGMD). Please note that the registration to CGAR does not automatically grant access to HGMD. Users need to obtain license from HGMD, separately.
The Human Gene Mutation Database (HGMD®) maintains a collection of known (published) genes associated with human inherited disease. HGMD® categorizes its data based on level of association with disease. Among those, variants with the most potential clinical implication would be:
- Disease causing mutation? (DM?): variant reported to be disease-causing in the report, yet author indicated some degree of doubt, or subsequence evidence caused the deleteriousness of the variant into question.
- Disease causing mutation (DM): pathological variant reported to be disease-causing.
HGMD tab in CGAR shows which variants in the current sample have matches from HGMD®.
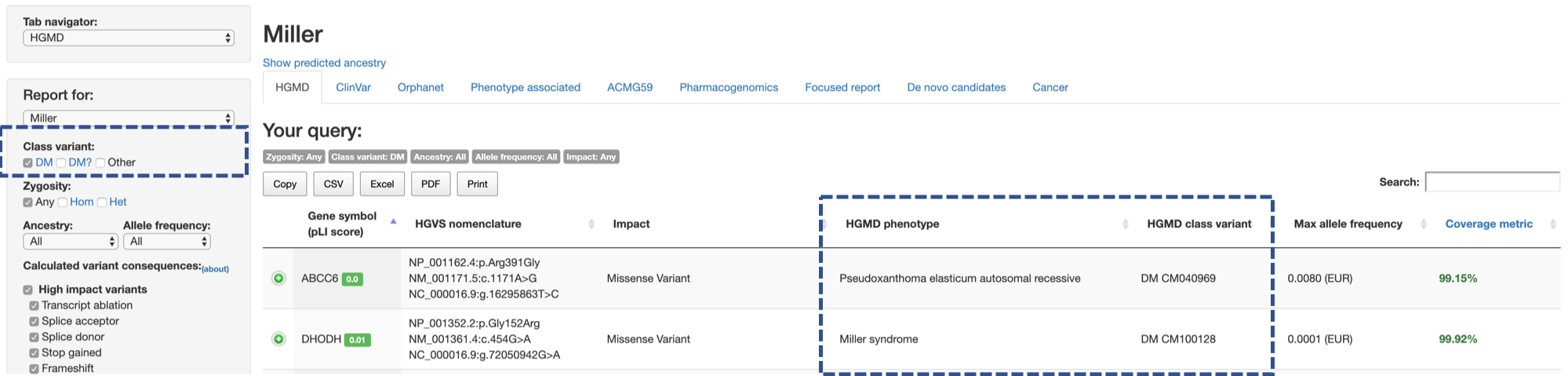
The Class variant on the side menu controls matches to which categories in HGMD will be shown (multiple choices are allowed).
- DM: show matches to DM variants.
- DM?: show matches to DM? variants.
- Other: show matches to other categories than DM or DM?.
By default, HGMD tab shows only variants that are categorized as DM in HGMD®, regardless of variant genotype, allele frequency, or variant consequences.
Zygosity: AnyClass variant: DMAncestry: AllAllele frequency: AllCalculated variant consequences: Any
For each variant, the main table on the right shows the disease phenotypes associated (or caused) by the variant (HGMD phenotype column) and categories and identifiers in HGMD® (HGMD class variant).