4.6. Putative phenotype-associated genes¶
For uses who want to focus on variants on genes associated to specific phenotypes or symptoms, but not yet certain about which genes to look for, CGAR provides a method to find candidate variants with phenotypes.
First, CGAR sends the free-form text input from user describing phenotypes to a machine-learning based text search engine in FindZebra.
Then, it retrieves information from a corpus of documents consisting of more than 36,000 entries from curated sources such as OMIM, Genetic and Rare Diseases Information Center, and OrphaNet, and ranks genes according to gene-specific scores for input phenotype.
From the ranked list of genes, CGAR selects the highest scored genes within user-specified proportion, and shows variants on the genes satisfying conditions on allele frequencies or consequences.
The advantage of using FindZebra is that users are not required to provide standardized ontology terms such as Systematized Nomenclature of Medicine - Clinical Terms (SNOMED CT) or Human Pathology Ontology (HPO).
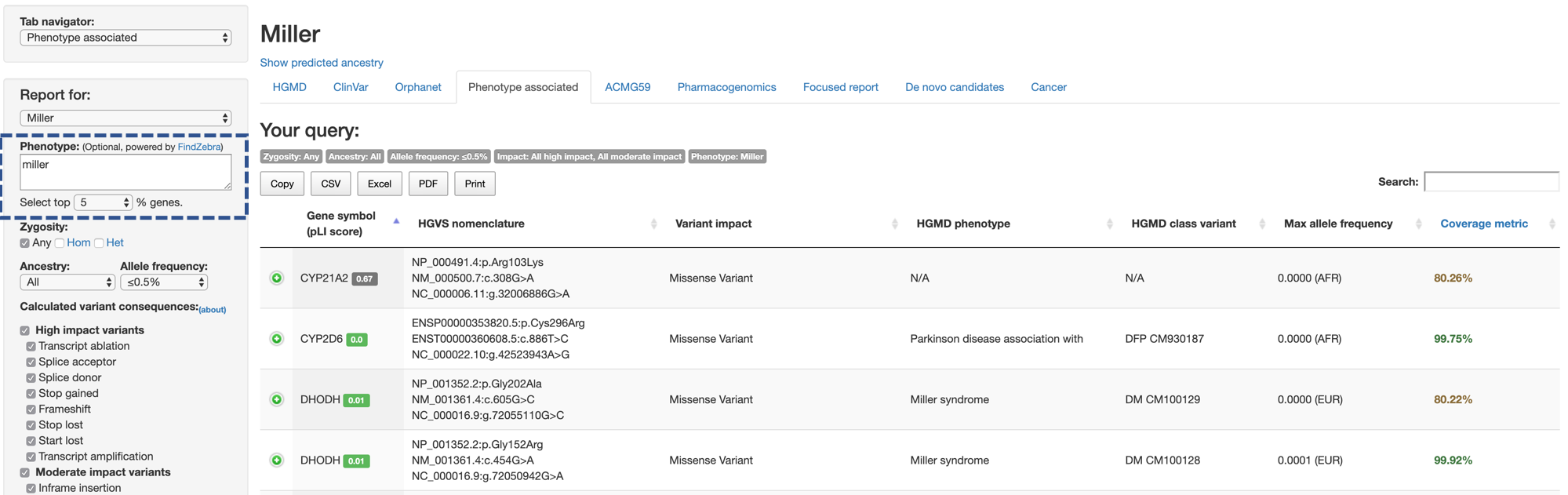
Here, users are only required to provide list of phenotypes (or descriptions) in the text box (Phenotype), and specify the percentage of high-scored genes you want to use from values 1 (use genes in top 1%), 5, 10, 25, 50, 100 (use all genes).