4.4. ClinVar variants¶
ClinVar is a freely accessible, public archive of reports of the relationships among human variations and phenotypes, with supporting evidence. ClinVar thus facilitates access to and communication about the relationships asserted between human variation and observed health status, and the history of that interpretation. (From ClinVar introduction)
ClinVar is a public database which archives reports on relationship between human genomic variants and phenotypes.
Like in HGMD tab, ClinVar tab in CGAR shows variants in the current sample that were previously submitted to ClinVar and their interpretations.
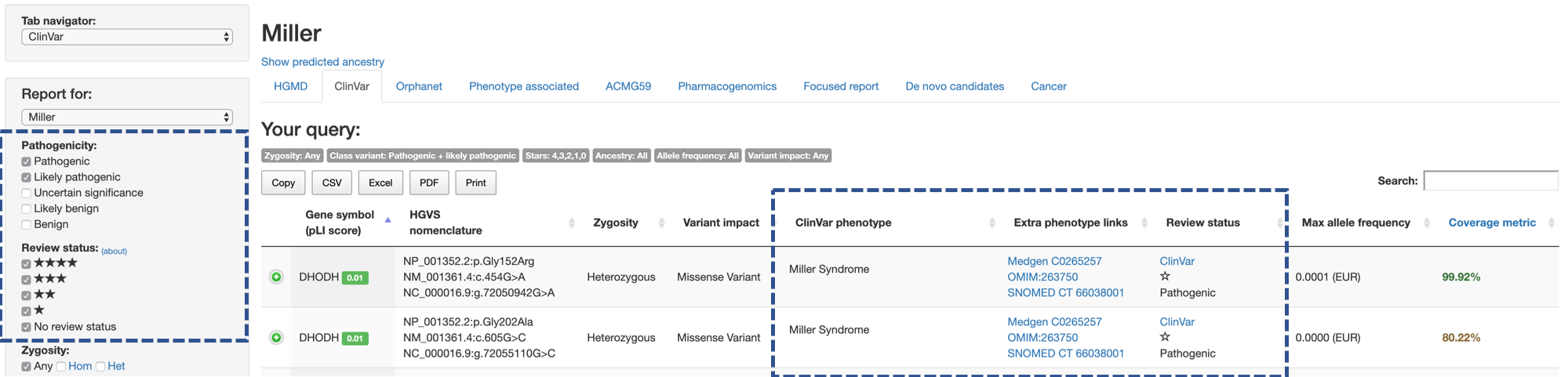
Users can filter variants by their reported clinical significance in ClinVar (Pathogenicity) and the level of review supporting the clinical significance (Review status).
In other words, CGAR can either be set to show variants which had been submitted to ClinVar with multiple evidence of its pathogenicity, or be set to show any variants which had ever been submitted to ClinVar - by controling Pathogenicity and Review status in combination of other controls in side menu.
Pathogenicitycan take any number of values from the 5 levels: Pathogenic (most deleterious) / Likely pathogenic / Uncertain significance / Likely benign / Benign (least deleterious).Review statusshows the number of gold starts which corresponds to each of the review statuses in ClinVar (link) (multiple values are allowed).
In the main table, CGAR shows the phenotype reported with the variant (ClinVar phenotype), links to phenotype databases (Extra phenotype links), and brief summary of variant’s status in ClinVar (Review status).
Extra phenotype linksprovide links to MedGen, Online Mendelian Inheritance in Men (OMIM), Systematized Nomenclature of Medicine - Clinical Terms (SNOMED CT), Human Pathology Ontology (HPO), and OrphaNet. If the variant is linked to multiple phenotypes, a set of links is provided for each phenotype.Review statuscolumn represents slightly different contents from side menu. In the main table, it contains:- Graphical representation of review status (stars)
- Aggregated interpretation of the variant in ClinVar
- Link to the variant’s record in Clinvar, where all submissions to ClinVar regarding the variant can be viewed.
Note
CGAR tries to maintain the most up-to-date data from all resources. However, there can be some discrepancy between what is available in ClinVAR and CGAR.