4.8. Pharmacogenomic variants¶
The Pharmacogenomics tab in CGAR reports variants or genes that could modulate drug metabolism or interfere with responses to drugs, summarized as in the following three categories:
- Variants specified in the dosage guidelines by the Clinical Pharmacogenetics Implementation Consortium (CPIC).
CPIC publishes guidelines to help clinicians to optimize drug therapy using genetic test results. The guidelines are published for pairs of drugs and genes, detailing drug therapy recommendations based on genotypes. CGAR lists all variants that were used in any of the guidelines, as a reminder to users (especially, clinicians) that some adjustment on drug prescription may be required.
Caution
The guidelines are based on allele types of genes, which may be defined by combination of genotypes in multiple loci. CGAR currently does not take multiple variants into consideration, and depending on quality of variant data, genotype of some loci might not be available, preventing the accurate application of guidelines. Thus, CGAR results in this section is best regarded as a reminder that some adjustments may be required. The actual adjustment should be dune after careful evaluation of alleles of the gene.
- Variants on genes from curated collection of genes Very Important Pharmacogene (VIP), playing important roles in drug metabolism or response.
The Pharmacogenomics Knowledge Base (PharmGKB) is a knowledge resource for the impact of human genetic variation on drug responses. One of most interesting feature in PharmGKB is the list of Very Important Pharmacogenes (VIPs), genes involved in metabolism of or response to drug(s). Thus, variants on these genes could have pharmacogenomic impact.
- Variants on genes selected for the investigation of drug response and genetic variation by the Phamacogenomics Research Network (PGRN).
PGRN developed a custom-capture panel of 84 genes associated with pharmacophenotypes, PGRNseq. Along with VIPs, these genes could be used to identify variants of pharmacogenomic impact.
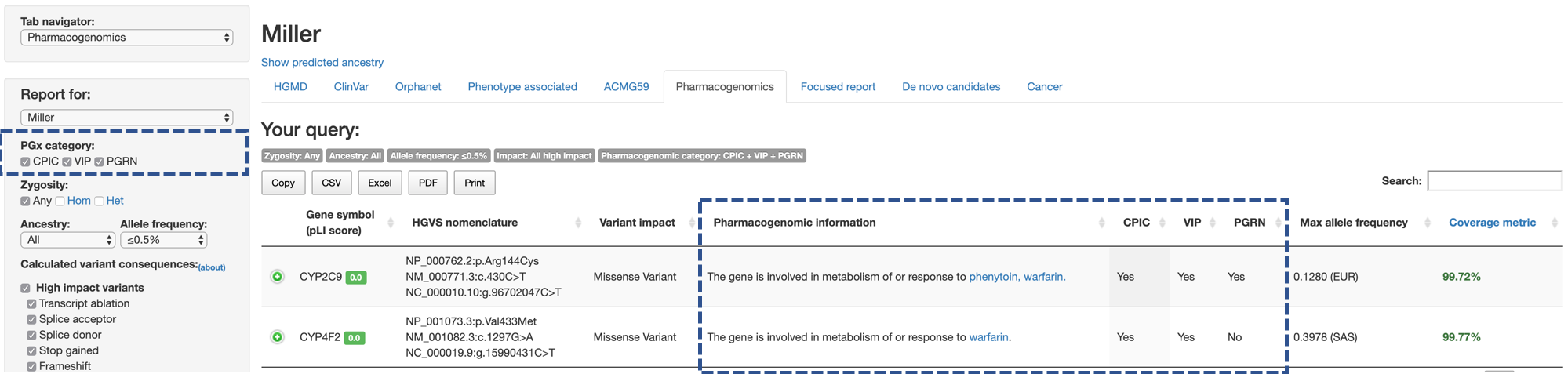
The PGx category on the side menu allows users to select which set of variants or genes would be used to identify variants with potential pharmacogenomic impact (multiple choices are allowed).
- CPIC: shows variants that matches to the variants in the CPIC guidelines.
- VIP: shows variants on genes listed as VIPs.
- PGRN: shows variants on genes included in PGRNseq panel.
By default, CGAR is set to use all of the above. For VIP and PGRN, rare and high impact variants are selected.
Zygosity: AnyAncestry: AllAllele frequency: <0.5%Calculated variant consequences: High impact variantsAllele frequency based on: Max
The columns CPIC, VIP, and PGRN in the main table shows the inclusion criteria for the variant.
For variants used in CPIC guideline, Phamacogenomic information columns shows list of drugs along with links to the CPIC guidelines.
For variants on genes in VIPs or PGRNseq, users are recommended to check repective documents.